Assembly theory
Assembly theory is a framework for quantifying selection and evolution. When applied to molecule complexity, its authors show it to be the first technique that is experimentally verifiable, unlike other molecular complexity algorithms that lack experimental measures.[1]
Background
The hypothesis was proposed by chemist Leroy Cronin in 2017 and developed by the team he leads at the University of Glasgow,[2][3] then extended in collaboration with a team at Arizona State University led by astrobiologist Sara Imari Walker, in a paper released in 2021.[4]
Assembly Theory conceptualizes objects not as point particles, but as entities defined by their possible formation histories.[5] This allows objects to show evidence of selection, within well-defined boundaries of individuals or selected units.[5] Combinatorial objects are important in chemistry, biology and technology, in which most objects of interest (if not all) are hierarchical modular structures.[5] For any object an 'assembly space' can be defined as all recursively assembled pathways that produce this object.[5] The 'assembly index' is the number of steps on a shortest path producing the object.[5] For such shortest path, the assembly space captures the minimal memory, in terms of the minimal number of operations necessary to construct an object based on objects that could have existed in its past.[5] The assembly is defined as "the total amount of selection necessary to produce an ensemble of observed objects"; for an ensemble containing objects in total, of which are unique, the assembly is defined to be
,
where denotes 'copy number', the number of occurrences of objects of type having assembly index .[5]
For example, the word 'abracadabra' contains 5 unique letters (a, b, c, d and r) and is 11 symbols long. It can be assembled from its constituents as a + b --> ab + r --> abr + a --> abra + c --> abrac + a --> abraca + d --> abracad + abra --> abracadabra, because 'abra' was already constructed at an earlier stage. Because this requires at least 7 steps, the assembly index is 7.[6] The word ‘abracadrbaa’, of the same length, for example, has no repeats so has an assembly index of 10.
Take two binary strings and as another example. Both have the same length bits, both have the same Hamming weight . However, the assembly index of the first string is ("01" is assembled, joined with itself into "0101", and joined again with "0101" taken from the assembly pool), while the assembly index of the second string is , since in this case only "01" can be taken from the assembly pool.[citation needed]
In general, for K subunits of an object O the assembly index is bounded by .[3]
Once a pathway to assemble an object is discovered, the object can be reproduced. The rate of discovery of new objects can be defined by the expansion rate , introducing a discovery timescale . [5] To include copy number in the dynamics of assembly theory, a production timescale is defined, where is the production rate of a specific object .[5] Defining these two distinct timescales , for the initial discovery of an object, and , for making copies of existing objects, allows to determine the regimes in which selection is possible.[5]
While other approaches can provide a measure of complexity, the researchers claim that assembly theory's molecular assembly number is the first to be measurable experimentally. Molecules with a high assembly index are very unlikely to form abiotically, and the probability of abiotic formation goes down as the value of the assembly index increases.[5] The assembly index of a molecule can be obtained directly via spectroscopic methods.[5][1] This method could be implemented in a fragmentation tandem mass spectrometry instrument to search for biosignatures.[1]
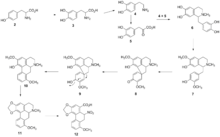
The theory was extended to map chemical space with molecular assembly trees, demonstrating the application of this approach in drug discovery,[2] in particular in research of new opiate-like molecules by connecting the "assembly pool elements through the same pattern in which they were disconnected from their parent compound(s)".
It is difficult to identify chemical signatures that are unique to life.[7] For example, the Viking lander biological experiments detected molecules that could be explained by either living or natural non-living processes.[8] It appears that only living samples can produce assembly index measurements above ~15.[1] However, 2021, Cronin first explained how polyoxometalates could have large assembly indexes >15 in theory due to autocatalysis. [9]
Critical views
Chemist Steven A. Benner has publicly criticized various aspects of Assembly Theory.[10] Benner argues that it is transparently false that non-living systems, and with no life intervention, cannot contain molecules that are complex but people would be misled in thinking that because it was published in Nature journals after peer review, these papers must be right.
A paper published in the Journal of Molecular Evolution[11] refers to Hector Zenil's blog post[12] "that identifies no less than eight fallacies of assembly theory". The paper also refers to the video essay by the same author[13] staying "that summarizes these fallacies, and highlights conceptual/methodological limitations, and the pervasive failure by the proponents of assembly theory to acknowledge relevant previous work in the field of complexity science". The paper concludes that "the hype around Assembly Theory reflects rather unfavorably both on the authors and the scientific publication system in general". The author[11] concludes that what "assembly theory really does is to detect and quantify bias caused by higher-level constraints in some well-defined rule-based worlds"; one "can use assembly theory to check whether something unexpected is going on in a very broad range of computational model worlds or universes".
The group led by Hector Zenil, a former Senior researcher and faculty member from Oxford and Cambridge and currently an Associate Professor in Biomedical Engineering from King's College London,[14] is cited to have reproduced the results of Assembly Theory with traditional statistical algorithms.[15]
Another paper authored by a group of chemists and planetary scientists, including an author affiliated with NASA, published in the journal of the Royal Society Interface[16] demonstrated that abiotic chemical processes have the potential to form crystal structures of great complexity — values exceeding the proposed abiotic/biotic divide of MA index = 15. They conclude that "while the proposal of a biosignature based on a molecular assembly index of 15 is an intriguing and testable concept, the contention that only life can generate molecular structures with MA index ≥ 15 is in error".
The paper also cites the papers and posts of Hector Zenil[17] as questioning whether a single scalar value like the assembly index can be employed to adequately discriminate between living and nonliving systems, and pointing out the noticeable similarities of the Assembly Theory approach to uncited prior efforts to distinguish biotic from abiotic molecular compounds.[18]
In particular, the paper mentions that Zenil and colleagues "may also have anticipated key conclusions of Assembly Theory by exploring connections among causal memory, selection, and evolution". [16] [19]
See also
References
- ^ a b c d Marshall SM, Mathis C, Carrick E, et al. (24 May 2021). "Identifying molecules as biosignatures with assembly theory and mass spectrometry". Nature Communications. 12 (3033): 3033. Bibcode:2021NatCo..12.3033M. doi:10.1038/s41467-021-23258-x. PMC 8144626. PMID 34031398.
- ^ a b Liu, Yu; Mathis, Cole; Bajczyk, Michał Dariusz; Marshall, Stuart M.; Wilbraham, Liam; Cronin, Leroy (2021). "Exploring and mapping chemical space with molecular assembly trees". Science Advances. 7 (39): eabj2465. Bibcode:2021SciA....7J2465L. doi:10.1126/sciadv.abj2465. PMC 8462901. PMID 34559562.
- ^ a b Marshall, Stuart M.; Murray, Alastair R. G.; Cronin, Leroy (2017). "A probabilistic framework for identifying biosignatures using Pathway Complexity". Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences. 375 (2109). arXiv:1705.03460. Bibcode:2017RSPTA.37560342M. doi:10.1098/rsta.2016.0342. PMC 5686400. PMID 29133442.
- ^ Sara Imari Walker; Leroy Cronin; Alexa Drew; Shawn Domagal-Goldman; Theresa Fisher; Michael Line; Camerian Millsaps (7 April 2019). "Probabilistic Biosignature Frameworks". In Victoria Meadows; Giada Arney; Britney Schmidt; David J. Des Marais (eds.). Planetary Astrobiology. doi:10.2458/azu_uapress_9780816540068-ch018.
- ^ a b c d e f g h i j k l Sharma, Abhishek; Czégel, Dániel; Lachmann, Michael; Kempes, Christopher P.; Walker, Sara I.; Cronin, Leroy (October 2023). "Assembly theory explains and quantifies selection and evolution". Nature. 622 (7982): 321–328. Bibcode:2023Natur.622..321S. doi:10.1038/s41586-023-06600-9. ISSN 1476-4687. PMC 10567559. PMID 37794189.
- ^ Mathis, Cole; Y. Patarroyo, Keith; Cronin, Lee. "Understanding Assembly Indices". Molecular Assembly. Cronin Group. Retrieved 26 March 2024.
resulting in an Assembly Index of 7
- ^ Schwieterman, Edward W.; Kiang, Nancy Y.; Parenteau, Mary N.; Harman, Chester E.; Dassarma, Shiladitya; Fisher, Theresa M.; Arney, Giada N.; Hartnett, Hilairy E.; Reinhard, Christopher T.; Olson, Stephanie L.; Meadows, Victoria S.; Cockell, Charles S.; Walker, Sara I.; Grenfell, John Lee; Hegde, Siddharth; Rugheimer, Sarah; Hu, Renyu; Lyons, Timothy W. (2018). "Exoplanet Biosignatures: A Review of Remotely Detectable Signs of Life". Astrobiology. 18 (6): 663–708. arXiv:1705.05791. Bibcode:2018AsBio..18..663S. doi:10.1089/ast.2017.1729. PMC 6016574. PMID 29727196.
- ^ Plaxco KW, Gross M (12 August 2011). Astrobiology: A Brief Introduction. JHU Press. pp. 285–286. ISBN 978-1-4214-0194-2. Retrieved 16 July 2013.
- ^ Cronin, Leroy (2021). "Exploring the Hidden Constraints that Control the Self-Assembly of Nanomolecular Inorganic Clusters". Bulletin of Japan Society of Coordination Chemistry. 78: 11–17. doi:10.4019/bjscc.78.11.
- ^ Benner, Steven A. "Assembly Theory and Agnostic Life Finding – The Primordial Scoop". Retrieved 19 September 2023.
- ^ a b Jaeger, Johannes (2024). "Assembly Theory: What It Does and What It Does Not Do". Journal of Molecular Evolution. 92 (2): 87–92. doi:10.1007/s00239-024-10163-2. PMC 10978598. PMID 38453740.
- ^ Zenil, Hector. "The 8 Fallacies of Assembly Theory – Medium". Retrieved 26 September 2023.
- ^ "Lee Cronin's Assembly Theory Disputed & Debunked by Dr. Hector Zenil - YouTube". YouTube. January 2024. Retrieved 13 March 2024.
- ^ "Dr Hector Zenil". Retrieved 17 March 2024.
- ^ Uthamacumaran, Abicumaran; Abrahão, Felipe S.; Kiani, Narsis; Zenil, Hector (2022). "On the Salient Limitations of the Methods of Assembly Theory and their Classification of Molecular Biosignatures". arXiv:2210.00901 [cs.IT].
- ^ a b Hazen, Robert M.; Burns, Peter C.; Cleaves II, H. James; Downs, Robert T.; Krivovichev, Sergey V.; Wong, Michael L. (2024). "Molecular assembly indices of mineral heteropolyanions: some abiotic molecules are as complex as large biomolecules". Journal of the Royal Society Interface. 21 (211). doi:10.1098/rsif.2023.0632. PMC 10878807. PMID 38378136.
- ^ Zenil, Hector. "The 8 Fallacies of Assembly Theory – Medium". Retrieved 26 September 2023.
- ^ Zenil, Hector; Kiani, Narsis A.; Shang, M-M; Tegnér, Jesper (2018). "Algorithmic complexity and reprogrammability of chemical structure networks". Parallel Processing Letters. 28 (1850005). arXiv:1802.05856. doi:10.1142/S0129626418500056.
- ^ Hernández-Orozco, Santiago; Kiani, Narsis A.; Zenil, Hector (2018). "Algorithmically probable mutations reproduce aspects of evolution, such as convergence rate, genetic memory and modularity". Royal Society Open Science. 5 (8). arXiv:1709.00268. doi:10.1098/rsos.180399. PMID 30225028.
Further reading
- Cronin, Leroy; Walker, Sara Imari (3 June 2016). "Beyond prebiotic chemistry". Science. 352 (6290): 1174–1175. Bibcode:2016Sci...352.1174C. doi:10.1126/science.aaf6310. ISSN 0036-8075. PMID 27257242. S2CID 206649123.
- Cronin, Leroy; Krasnogor, Natalio; Davis, Benjamin G.; Alexander, Cameron; Robertson, Neil; Steinke, Joachim H. G.; Schroeder, Sven L. M.; Khlobystov, Andrei N.; Cooper, Geoff; Gardner, Paul M.; Siepmann, Peter (2006). "The imitation game—a computational chemical approach to recognizing life". Nature Biotechnology. 24 (10): 1203–1206. doi:10.1038/nbt1006-1203. ISSN 1546-1696. PMID 17033651. S2CID 4664573.
- Ball, Philip (4 May 2023). "A New Theory for the Assembly of Life in the Universe". Quanta Magazine.








![{\displaystyle C=[01010101]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/42a88f4161802acd454b1d3ffc767325cd1fdc80)
![{\displaystyle D=[00010111]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/846a858cf172fd15e4382568da0c30d3e568fc88)











