Vts1: Difference between revisions
Content deleted Content added
m Open access bot: doi added to citation with #oabot. |
m Normalize {{Multiple issues}}: Remove {{Multiple issues}} for only 1 maintenance template(s): Technical |
||
| Line 1: | Line 1: | ||
{{technical|date=August 2016}} |
|||
{{Orphan|date=November 2014 |
{{Orphan|date=November 2014}} |
||
{{Infobox protein family |
{{Infobox protein family |
||
Revision as of 19:14, 31 May 2020
This article may be too technical for most readers to understand. (August 2016) |
| Vts1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of Vts1p–SRE complex.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Vts1 | ||||||||
| Pfam | PF07647 | ||||||||
| SCOP2 | 1b0x / SCOPe / SUPFAM | ||||||||
| |||||||||
Vts1 is a post-transcriptional regulator that has RNA-binding Sterile alpha motif (SAM) domain.[2][3] The protein is found in Saccharomyces cerevisiae and several eukaryotes. In Saccharomyces the Vts1 impacts vesicular transport and sporulation.[4][5]
Interactions
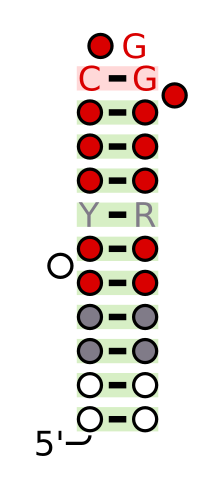
Protein-protein interactions through SAM domains participate in different regulatory activities such as signal transduction. Proteins having such domains were also shown to recognize and interact with RNA structures of similar shape to the Smaug response element (SRE).[6] Vts1 binds to RNA targets that have CUGGC on hairpin loops.[7]
References
- ^ Philip E Johnson; Logan W Donaldson (Jan 2006). "SRNA recognition by the Vts1p SAM domain". Nature Structural & Molecular Biology. 13 (1): 177–178. doi:10.1038/nsmb1039.
{{cite journal}}: Unknown parameter|lastauthoramp=ignored (|name-list-style=suggested) (help) - ^ Bork P, Ponting CP, Hofmann K, Schultz J (1997). "SAM as a protein interaction domain involved in developmental regulation". Protein Sci. 6 (1): 249–253. doi:10.1002/pro.5560060128. PMC 2143507. PMID 9007998.
- ^ Florian C Oberstrass, Albert Lee, Richard Stefl, Michael Janis, Guillaume Chanfreau & Frédéric H-T Allain (Jan 2006). "Shape-specific recognition in the structure of the Vts1p SAM domain with RNA". Nature Structural & Molecular Biology. 13: 160–167. doi:10.1038/nsmb1038. PMID 16429156.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Adam M. Deutschbauer, Roy M. Williams, Angela M. Chu, and Ronald W. Davis (November 2002). "Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyce scerevisiae". PNAS. 99 (24): 15530–15535. doi:10.1073/pnas.202604399. PMC 137751. PMID 12432101.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Meik Dilcher, Beate Köhler and Gabriele Fischer von Mollard (September 2001). "Genetic Interactions with the Yeast Q-SNARE VTI1 Reveal Novel Functions for the R-SNARE YKT6". The Journal of Biological Chemistry. 276: 34537–34544. doi:10.1074/jbc.M101551200. PMID 11445562.
- ^ Traci M Tanaka Hall (2003). "SAM breaks its stereotype" (PDF). Nature Structural & Molecular Biology. 10: 677–679. doi:10.1038/nsb0903-677.
- ^ Florian C Oberstrass, Albert Lee, Richard Stefl, Michael Janis, Guillaume Chanfreau and Frédéric H-T Allain (January 2006). "Shape-specific recognition in the structure of the Vts1p SAM domain with RNA". Nature Structural & Molecular Biology. 13 (2): 160–167. doi:10.1038/nsmb1038. PMID 16429156.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
