STK3
Serine/threonine-protein kinase 3 is an enzyme that in humans is encoded by the STK3 gene.[5][6]
Background[edit]
Protein kinase activation is a frequent response of cells to treatment with growth factors, chemicals, heat shock, or apoptosis-inducing agents. This protein kinase activation presumably allows cells to resist unfavorable environmental conditions. The yeast 'sterile 20' (Ste20) kinase acts upstream of the mitogen-activated protein kinase (MAPK) cascade that is activated under a variety of stress conditions. MST2 was first identified as a kinase that resembles budding yeast Ste20 (Creasy and Chernoff, 1996) and later as a kinase that is activated by the proapoptotic agents straurosporine and FAS ligand (MIM 134638) (Taylor et al., 1996; Lee et al., 2001).[supplied by OMIM][6]
Structure[edit]
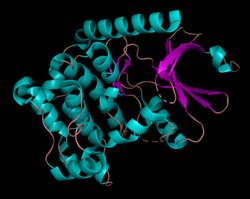
Human serine/threonine-protein kinase 3 (STK3, or MST2) is a 56,301 Da[7] monomer with three domains: a SARAH domain, composed of a long α-helix at the C-terminus that when dimerized, forms an antiparallel dimeric coiled-coil, an inhibitory domain, and a catalytic kinase domain at the N-terminus.[8] The SARAH (Salvador/RASSF/Hpo) domain has been found to mediate dimeric interactions between MST2 and RASSF enzymes, a class of tumor suppressors that serve an important role in activating apoptosis, as well as between MST2 and SAV1, a non-catalytic polypeptide responsible for bringing MST2 to an apoptotic pathway.[9][10] When the MST2 kinase domain is in its active state, a threonine residue residing on an alpha helix at the 180th position (T180) is autophosphorylated.[11]

Mechanism[edit]
Activation[edit]
STK3 is activated through autophosphorylation by dimerizing with itself or heterodimerizing with its homolog, MST1 (STK4).[12] Heterodimerization has been shown to exhibit a roughly six-fold weaker binding affinity than homodimerization with MST2, as well as lower kinase activity compared to both MST2/MST2 and MST1/MST1 homodimers.[10] In addition to activation by straurosporine and FAS ligand, STK3 has been found to be activated through dissociation of GLRX and Thioredoxin (Trx1) from STK3 under oxidative stress.[12] Recent studies have shown that when caspase 3 is activated during apoptosis, MST2 is cleaved, resulting in removal of the regulatory SARAH and inhibitory domains and thus regulation of MST2's kinase activity. Because cleavage by caspase 3 also cleaves off MST2's nuclear export signal, the MST2 kinase fragment can diffuse into the nucleus and phosphorylate Ser14 of histone H2B, promoting apoptosis.[10]
Inactivation[edit]
Inactivation of MST2 can be accomplished in several ways, including inhibition of MST2 homodimerization and autophosphorylation by c-Raf, which binds to the MST2 SARAH domain,[10] and phosphorylation of the highly conserved Thr117 by Akt (protein kinase B), blocking autophosphorylation of Thr180, MST2 cleavage, kinase activity, and translocation to the nucleus.[13]
MST2 substrates[edit]
In the mammalian Hippo signaling pathway, MST2, along with its homolog MST1, serves as an upstream kinase whose catalytic activity is responsible for downstream events leading to downregulation of proliferation-associated genes and increased transcription of proapoptotic genes.[12] When MST2 binds to SAV1 through its SARAH domain, MST2 phosphorylates LATS1/LATS2 with the help of SAV1, MOB1A/MOB1B, and Merlin (protein). In turn, LATS1/LATS2 phosphorylates and inhibits YAP1, preventing its movement into the nucleus and activation of transcription of pro-proliferative, anti-apoptotic and migration-associated genes. In the cytoplasm, YAP1 is marked for degradation by the SCF complex.[14] Additionally, MST2 phosphorylates transcription factors in the FOXO (Forkhead box O) family, which diffuse into the nucleus and activate transcription of pro-apoptotic genes.[12]
Disease Relevance[edit]
In many types of cancers, the proto-oncogene c-Raf binds to the SARAH domain of MST2 and prevents RASSF1A-mediated MST2 dimerization and subsequent downstream pro-apoptotic signaling.[15] Research has shown that in cells with loss of PTEN (gene), a tumor suppressor that is frequently mutated in cancers, Akt activity is upregulated, resulting in increased MST2 inactivation and undesirable cell proliferation.[16]
References[edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000104375 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000022329 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Taylor LK, Wang HC, Erikson RL (September 1996). "Newly identified stress-responsive protein kinases, Krs-1 and Krs-2". Proceedings of the National Academy of Sciences of the United States of America. 93 (19): 10099–104. Bibcode:1996PNAS...9310099T. doi:10.1073/pnas.93.19.10099. PMC 38343. PMID 8816758.
- ^ a b "Entrez Gene: STK3 serine/threonine kinase 3 (STE20 homolog, yeast)".
- ^ "PhosphoSitePlus: Serine/threonine-protein kinase 3 - Protein Information".
- ^ Liu G, Shi Z, Jiao S, Zhang Z, Wang W, Chen C, Hao Q, Hao Q, Zhang M, Feng M, Xu L, Zhang Z, Zhou Z, Zhang M (March 2014). "Structure of MST2 SARAH domain provides insights into its interaction with RAPL". Journal of Structural Biology. 185 (3): 366–74. doi:10.1016/j.jsb.2014.01.008. PMID 24468289.
- ^ Sánchez-Sanz G, Tywoniuk B, Matallanas D, Romano D, Nguyen LK, Kholodenko BN, Rosta E, Kolch W, Buchete NV (October 2016). "SARAH Domain-Mediated MST2-RASSF Dimeric Interactions". PLOS Computational Biology. 12 (10): e1005051. Bibcode:2016PLSCB..12E5051S. doi:10.1371/journal.pcbi.1005051. PMC 5055338. PMID 27716844.
- ^ a b c d Galan JA, Avruch J (Sep 2016). "MST1/MST2 Protein Kinases: Regulation and Physiologic Roles". Biochemistry. 55 (39): 5507–5519. doi:10.1021/acs.biochem.6b00763. PMC 5479320. PMID 27618557.
- ^ Ni L, et al. (Oct 2013). "Structural Basis for Autoactivation of Human Mst2 Kinase and Its Regulation by RASSF5". Structure. 21 (10): 1757–1768. doi:10.1016/j.str.2013.07.008. PMC 3797246. PMID 23972470.
- ^ a b c d Lessard-Beaudoin M, Laroche M, Loudghi A, Demers MJ, Denault JB, Grenier G, Riechers SP, Wanker EE, Graham RK (November 2016). "Organ-specific alteration in caspase expression and STK3 proteolysis during the aging process" (PDF). Neurobiology of Aging. 47: 50–62. doi:10.1016/j.neurobiolaging.2016.07.003. PMID 27552481. S2CID 3930860.
- ^ Kim D, Shu S, Coppola MD, Kaneko S, Yuan Z, Cheng JQ (Mar 2010). "Regulation of Proapoptotic Mammalian ste20–Like Kinase MST2 by the IGF1-Akt Pathway". PLOS ONE. 5 (3): e9616. Bibcode:2010PLoSO...5.9616K. doi:10.1371/journal.pone.0009616. PMC 2834758. PMID 20231902.
- ^ Meng Z, Moroishi T, Guan K (Jan 2016). "Mechanisms of Hippo pathway regulation". Genes Dev. 30 (1): 1–17. doi:10.1101/gad.274027.115. PMC 4701972. PMID 26728553.
- ^ Nguyen LK, Matallanas DG, Romano D, Kholodenko BN, Kolch W (Jan 2015). "Competing to coordinate cell fate decisions: the MST2-Raf-1 signaling device". Cell Cycle. 14 (2): 189–199. doi:10.4161/15384101.2014.973743. PMC 4353221. PMID 25607644.
- ^ Romano D, Matallanas D, Weitsman G, Preisinger C, Ng T, Kolch W (Feb 2010). "Proapoptotic kinase MST2 coordinates signaling crosstalk between RASSF1A, Raf-1, and Akt". Cancer Res. 70 (3): 1195–1203. doi:10.1158/0008-5472.CAN-09-3147. PMC 2880716. PMID 20086174.
Further reading[edit]
- Creasy CL, Chernoff J (September 1995). "Cloning and characterization of a human protein kinase with homology to Ste20". The Journal of Biological Chemistry. 270 (37): 21695–700. doi:10.1074/jbc.270.37.21695. PMID 7665586.
- Schultz SJ, Nigg EA (October 1993). "Identification of 21 novel human protein kinases, including 3 members of a family related to the cell cycle regulator nimA of Aspergillus nidulans". Cell Growth & Differentiation. 4 (10): 821–30. PMID 8274451.
- Creasy CL, Chernoff J (December 1995). "Cloning and characterization of a member of the MST subfamily of Ste20-like kinases". Gene. 167 (1–2): 303–6. doi:10.1016/0378-1119(95)00653-2. PMID 8566796.
- Bren A, Welch M, Blat Y, Eisenbach M (September 1996). "Signal termination in bacterial chemotaxis: CheZ mediates dephosphorylation of free rather than switch-bound CheY". Proceedings of the National Academy of Sciences of the United States of America. 93 (19): 10090–3. Bibcode:1996PNAS...9310090B. doi:10.1073/pnas.93.19.10090. PMC 38341. PMID 8816756.
- Wang HC, Fecteau KA (August 2000). "Detection of a novel quiescence-dependent protein kinase". The Journal of Biological Chemistry. 275 (33): 25850–7. doi:10.1074/jbc.M000818200. PMID 10840030.
- Lee KK, Ohyama T, Yajima N, Tsubuki S, Yonehara S (June 2001). "MST, a physiological caspase substrate, highly sensitizes apoptosis both upstream and downstream of caspase activation". The Journal of Biological Chemistry. 276 (22): 19276–85. doi:10.1074/jbc.M005109200. PMID 11278283.
- De Souza PM, Kankaanranta H, Michael A, Barnes PJ, Giembycz MA, Lindsay MA (May 2002). "Caspase-catalyzed cleavage and activation of Mst1 correlates with eosinophil but not neutrophil apoptosis". Blood. 99 (9): 3432–8. doi:10.1182/blood.V99.9.3432. PMID 11964314. S2CID 8728566.
- Deng Y, Pang A, Wang JH (April 2003). "Regulation of mammalian STE20-like kinase 2 (MST2) by protein phosphorylation/dephosphorylation and proteolysis". The Journal of Biological Chemistry. 278 (14): 11760–7. doi:10.1074/jbc.M211085200. PMID 12554736.
- Rabizadeh S, Xavier RJ, Ishiguro K, Bernabeortiz J, Lopez-Ilasaca M, Khokhlatchev A, Mollahan P, Pfeifer GP, Avruch J, Seed B (July 2004). "The scaffold protein CNK1 interacts with the tumor suppressor RASSF1A and augments RASSF1A-induced cell death". The Journal of Biological Chemistry. 279 (28): 29247–54. doi:10.1074/jbc.M401699200. PMID 15075335.
- O'Neill E, Rushworth L, Baccarini M, Kolch W (December 2004). "Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1". Science. 306 (5705): 2267–70. Bibcode:2004Sci...306.2267O. doi:10.1126/science.1103233. PMID 15618521. S2CID 30879956.
- Chan EH, Nousiainen M, Chalamalasetty RB, Schäfer A, Nigg EA, Silljé HH (March 2005). "The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1". Oncogene. 24 (12): 2076–86. doi:10.1038/sj.onc.1208445. PMID 15688006. S2CID 27285160.
- Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, Im CR, Lee JO, Yonehara S, Lim DS (March 2006). "Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis". Cancer Research. 66 (5): 2562–9. doi:10.1158/0008-5472.CAN-05-2951. PMID 16510573.
- Callus BA, Verhagen AM, Vaux DL (September 2006). "Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation". The FEBS Journal. 273 (18): 4264–76. doi:10.1111/j.1742-4658.2006.05427.x. PMID 16930133. S2CID 8261982.
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (November 2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983. S2CID 7827573.
- Seidel C, Schagdarsurengin U, Blümke K, Würl P, Pfeifer GP, Hauptmann S, Taubert H, Dammann R (October 2007). "Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma". Molecular Carcinogenesis. 46 (10): 865–71. doi:10.1002/mc.20317. PMID 17538946. S2CID 36848574.
- Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O'neill E (September 2007). "RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein". Molecular Cell. 27 (6): 962–75. doi:10.1016/j.molcel.2007.08.008. PMC 2821687. PMID 17889669.